Table of Contents
Proteins
Introduction to proteins
Proteins are linear chains of amino acids, typically several hundred units long. There are 20 standard amino acids in biology, each with a unique residue. The behavior of a protein is defined by the sequence of residues. Some are hydrophobic, some are hydrophillic, some have positive charge at pH 7, others have negative charge. There are free online databases to look up the sequence of any protein. For many proteins the folded structure(s) is also known from x-ray crystallography data (see the protein databank).
A single protein may exist in several different structures. The motion between structures is an important aspect of its biological function. There is a molecular morphing database that provide moives showing the approximate motion between different strucutres.
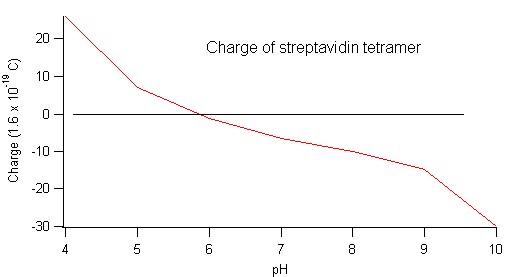
Example: Estimate the size, charge and weight of streptavidin. Start with an overview by reading wikipedia. Then follow the link to the protein database.
I search for streptavidin and pick pdb# 2IZI. In the pdb file header there is sequence of 123 residues (the core of the protein):
- SEQRES 1 A 123 ALA GLU ALA GLY ILE THR GLY THR TRP TYR ASN GLN LEU
- SEQRES 2 A 123 GLY SER THR PHE ILE VAL THR ALA GLY ALA ASP GLY ALA
- SEQRES 3 A 123 LEU THR GLY THR TYR GLU SER ALA VAL GLY ASN ALA GLU
- SEQRES 4 A 123 SER ARG TYR VAL LEU THR GLY ARG TYR ASP SER ALA PRO
- SEQRES 5 A 123 ALA THR ASP GLY SER GLY THR ALA LEU GLY TRP THR VAL
- SEQRES 6 A 123 ALA TRP LYS ASN ASN TYR ARG ASN ALA HIS SER ALA THR
- SEQRES 7 A 123 THR TRP SER GLY GLN TYR VAL GLY GLY ALA GLU ALA ARG
- SEQRES 8 A 123 ILE ASN THR GLN TRP LEU LEU THR SER GLY THR THR GLU
- SEQRES 9 A 123 ALA ASN ALA TRP LYS SER THR LEU VAL GLY HIS ASP THR
- SEQRES 10 A 123 PHE THR LYS VAL LYS PRO
Each 3 letter code stands for a different amino acid - i.e. a different residue (key).
I put the sequence into the protein calculator and submit a query asking for molecular mass, isoelectric point and charge at different pH values:
Example: Visualizing surface charge of protein using VMD
Example: tips about pH measurements
Protocols for handling protiens
Be careful not to contaminate a protein solution by touching the liquid, or anything that will contact the liquid. Our bodies exude enzymes that digest proteins. These enzymes are called proteases. It is a serious mistake to touch a pipette tip, or the inside of an lid, with your bare finger.
Protein are typically stored in eppendorf tubes. Proteins are never kept in glassware because the proteins stick to the glass surface.
Protein should always be concentrated and frozen for long term storage. A typical concentration is 1 mg/ml. When protein is needed for an experiment, one eppendorf tube is taken out of the deep freeze - the other “aloquots” are kept frozen.
To mix a protein solution, gently pipette in and out several times. Don't create any bubbles, because liquid-air interfaces will denature proteins. Never vortex mix a protein solution.
If protein is on the sidewalls of an eppendorf tube, briefly spin the tube in a centrifuge so that all protein is at the bottom of the eppendorf tube.
Software for analyzing protein structure
Much of this software was developed by protein crystallographers - Karplus group at OSU are experts in this field.
- VMD - Protein visualization program
- PDB2PQR - Compute each atom's charge in a protein
- PyMol - Protein visualization program with some manipulation capabilities
- Coot - Model building program designed to build protein structures to fit electron density maps acquired from crystallography experiments.
- O - Model building and refinement program for crystallography
- Emacs - Protein coordinate files contain lots of information and are difficult to work with in notepad. Emacs is a code writing software that allows one to make large & unusual modifications text files easily.
- Argus Lab - Make homemade molecular coordinate files in pdb format
Animations of protein function
Protein conjugation
Protein conjugation means covalent linkage between a protein and something else. Proteins have many amine groups on their surface due to the common protein residue lysine. These amine groups are the easiest target for conjugation. An amine-reactive group, such as succinimidyl ester (NHS), is used to forge a covalent bond onto the amine group of the protein. Biotech companies use many many types of “amine reactive probes”. These are fluorescent molecules that have already been prepared with an amine-reactive group.
Thiol reactive linker molecules are an alternative to amine-reactive linker molecules.
Surface functionalization of a nanotube
1-pyrenebutanoic acid, succinimidyl ester is an amine-reactive probe sold by Invitrogen (see - Labeling & Detection (Molecular Probes) - Reactive Dyes and Chemicals). Most people utilize the fluorescent properties of pyrene, but we utilize the specific affinity of pyrene to bind to a nanotube surface. A recipe for anchoring amine-reactive pyrene to a nanotube is given by Besteman et al, Nano Letters (2003). The next step, conjugation of a protein to the amine-reactive nanotube anchor, is detailed on the Invitrogen product page.
Surface functionalization of silicon oxide
A silicon oxide surface can be coated with amine groups and then further “lego blocks” can be added to make a protein functionalized surface.
To get amine termination on a silicon oxide surface, the most common procedure is exposure to APTES, the acronym stands for 3-aminopropyl trimethoxysilane (also abreiviated to APS) (Sigma Aldrich 09326-100ML), formula H2N(CH2)3Si(OCH3)3, 179.29 g/mol. Several group on campus are using this technique.
- Esha Chatterjee, grad student from Chemistry department (collaborates with Pallavi Dhagat), uses a simple proceedure: submerge the chip in 10% aqueous solution of APTES for about 2 hours at room temperature. Be aware that APTES has a very short half life in aqueous solution ~ 7 hours.
- Debbie Gale, grad student in Bioengineering, Rorrer group, uses a slightly different proceedure. The APTES is diluted in ethanol (50uL:2mL) and the chip is submerged for 2 hours at T = 80 C.
- Grad student in Bioengineering, Christine Kelly's group, anchors proteins onto the oxide surface using long tethers. The tethers can be designed to stick into solutions (eg. hydrophillic tethers will stick into the water).
There are many variation of the APTES functionalization proceedure in the biosensor literature, see Cui, Science 2001, Heath JACS 2007. etc.
For a cheap, easy proof-of-principle experiment, the “lego block” on the amine terminated silicon oxide could be amine-reactive biotin. Esha Chatterjee uses Sulfo-NHS-LC-biotin (Pierce Biotech, # 21335). It is water soluble and is capable of directly attaching to primary amine groups on the silicion oxide. Reaction instructions are available from the Pierce website. Debbie Gale, grad student from Bio-Engineering, uses EZ-Link Sulfo-NHS-Biotin (sulfosuccinimidobiotin) (Pierce Biotechnology, #21217). Another option is Sulfo-NHS biotin, it has no spacer (LC) and hence, can yield higher surface density of biotin molecules.
Surface passivication
The 2008 Nature Method's paper from TJ Ha's group includes online supplimentary info, like this video demonstration of PEG-ylation.
http://bio.physics.uiuc.edu/movies/PEG%20protocol/
These movies are also on the T: drive in the Manuals folder.
Fluorescence
| - | Excite (nm) | Emit (nm) | notes |
|---|---|---|---|
| Fluorescein | 494 | 521 | This is the dye in FITC, a functionalized version of fluorescein. DyLight488 works with the same filter set. |
| Cy3 | 550 | 570 | Commonly used by Indra lab, works with Oksana's 532 green laser. Molecule is equivalent to Dylight549 |
| Cy5 | 649 | 670 | emission is basically invisible to human eye. Molecule is equivalent to DyLight649 |
- The Zeiss widefield microscope in the pharmacy building has filter sets for all three exite/emit combos.
- The Zeiss widefiled microscope in Chemistry (Remcho lab) has the filter set for fluorescein.
Tips for dispersing single fluorophore molecules on a glass slide and visualizing.
Immunoassays
Immunoassays are inspired by the molecular regonition processes of the immune system. In the immune system, a foreign molecule is “suffocated” when antibody molecules bind onto it. In a typical immunoassay, the antibody molecule is conjugated to a surface and a lab technican checks to see if the antibody's target is present in a blood sample.
Examples of common “model systems” for immunoassays are
- The IgG molecule and its antibody (anti-IgG)
- Prostrate specific antigen (PSA) and it's antibody
Vince Remcho in the chemistry department has lots of experience with the PSA system. Debbie Gale in bioengineering is working with the IgG system. An even simpler test system is biotin-streptavidin (very high binding affinity). Pallavi Dhagat's group in EECE is using biotin-streptavidin for testing devices.
When one chip is used to test for many different proteins it is called an integrated immunoassays. Not all immunoassay technologies are capable of integration. Fluorescene labelling is currently the “gold standard”. Nano-FETs and magnetic bead sensors are promising candiates for faster and cheaper integrated immunoassays. In an integrated immunoassay, different probe molecules must be stuck to different parts of the chip. These protein arrays are created by printing spots of protein (for example, antibodies, recombinant proteins, peptides) on a solid surface such as a glass slide coated with a protein-binding chemistry. For such arrays, it is typical to modify the surface with molecules such as nickel or streptavidin to bind His-tagged or biotin-labeled proteins, respectively. This limits the range of proteins to be arrayed and requires specific surfaces for specific protein constructs. The production of proteins in bacterial culture systems are well-established procedures, and such proteins can be expressed with specific fusion tags for immobilization to appropriately coated solid surfaces. For nontagged proteins, the same surfaces used for DNA arrays such as amine, aldehyde or epoxy chemistries can be used for the capture of proteins onto the array.
Many companies make “spotters” for printing arrays of protiens (also called 'microarrayers'). Examples are listed below:
Diffusion
The Einstein-Stokes equation is a good method of estimating diffusion coefficients based on the size of the molecule.
For proteins in water the formula reduces to D = (200 um^2/s)/r where r is the Stokes radius of the protein measured in nm.
Here are a few numbers
- r = 0.1 nm (O2 molecule in water). D = 2000 um^2/s
- r = 0.4 nm (sugar molecule in water. D = 500 um^2/s
- r = 2 nm (small globular protein). D = 100 um^2/s
- r = 20 nm (very large protein or an extended polymer like dna). D = 10 um^2/s
So an oxygen molecule will take about 5 seconds to diffuse through 10 microns of water. However, to move 1 cm through water (by pure diffusion) it would take a full day. Cells (which are about 10 microns in size) are designed to work by diffusive transport on the time scale of seconds to minutes.
Lengths scales vs. Concentration
| Concentration | Number density | Avg separation |
|---|---|---|
| 10 mM | 6e24 per cubic meter | 5 nm |
| 10 uM | 6e21 per cubic meter | 50 nm |
| 10 nM | 6e18 per cubic meter | 500 nm |
Electrostatics
“Well known cases in which electrostatics are important are the protein-ligand attractive electrostatic interactions shown for superoxide dismutases, the trypsin-BPTI complex, the barstar-barnase system and DNA binding proteins.” Nielsen et al. Prot. Eng. (1999).
Cell Lysates
Standard protocol for extracting the proteins from cells.